Tweedue and Recipes part 2 (kaggle data)
I just got my feet wet with tweedie regression and the recipes package yesterday. The results have been underwhelming, as the models didnt appear that predictive. I figured I might give it another try, this time using Kaggle’s claim prediction challenge from 2012.
It is no longer possible to submit models, so we will create our own 20% test sample from the kaggle training data set and see how we fare. Submissions were evaluated on the Normalized Gini coefficient.
Overview of the approach:
We split the train data (13M obs, 35 columns) into 80% train and 20% test. We will use k-fold cross validation to find the best number of iterations for an xgboost tweedie model using all available columns but no feature engineering. We will train a new model on the full 80% training dataset. The 20% test dataset will then be scored and the model prediction will be evaluated using normalized gini, as was done in the competition.
I don’t do anything clever such as feature engineering or capping individual losses, so I dont expect a very good leaderboard performance. That being said, this is a very old competition from 2012, so they didnt have access to xgboost… let’s see!
My 32GB of RAM was a bit short to work with the data. I saved a lot to disk, removing objects that weren’t absolutely necessary.
Corners were slightly cut, such as:
- The prep() function that trains all the knn imputation models was only run on a 1e5 of the 10M records.
- The k-fold cross validation only had 3 folds and was performed on 3e6 observations. The best iteration was 310.
- The full model was trained on 6e6 observations (instead of 10e6).
Libraries
The usual tidyverse suspects are used for data wrangling and plots. I use themes and palettes from Claus Wilke’s dviz.supp and colorblindr packages. Plots in the lift charts are arranged in a grid using the patchwork package.
The pre-processing pipeline uses recipes, rsample and yardstick.
Vroom is used to read the csv. fstis used to save and read intermediate outputs.
MLmetricsis used to compute the normalised gini coefficient.
library(tidyverse) # used for data wrangling
library(xgboost) # used for modelling
library(tictoc) # to time execution
library(dviz.supp) # devtools::install_github("clauswilke/dviz.supp")
library(colorblindr) # devtools::install_github("clauswilke/colorblindr")
library(patchwork) # to arrange plots in a grid
library(rsample) # create train/test sample
library(yardstick)
library(recipes)
library(vroom) # for quick reading of csv
library(fst) # for quick saving of tables
library(MLmetrics) # for normalized gini # kaggle claim data from https://www.kaggle.com/c/ClaimPredictionChallenge/daatre
if(FALSE){
kaggle_train <- vroom::vroom(here::here("content/post/data/downloads/train_set.csv")) %>%
select(-Row_ID, -Household_ID, -Vehicle)
set.seed(42)
#https://cran.r-project.org/web/packages/recipes/vignettes/Simple_Example.html
split <- rsample::initial_split(kaggle_train, prop = 0.8)
train <- rsample::training(split)
test <- rsample::testing(split)
train_1e5_split <- rsample::initial_split(train, prop = 0.01) # 1e5 obs used for prepping recipe
train_1e5 <- rsample::training(train_1e5_split)
fst::write_fst(train, here::here("content/post/data/interim/claims_train.fst"))
fst::write_fst(train_1e5, here::here("content/post/data/interim/claims_train_1e5.fst"))
fst::write_fst(test, here::here("content/post/data/interim/claims_test.fst"))
rm(list=ls()) # clear memory of object but keep libraries.
invisible(gc())
}Prepare recipe
Here we only load the train dataset because of memory issues.
step_string2factor(all_nominal()) is required because step_knnimpute() doesnt work with character columns.
I used head(1e4) to “train” the recipe because otherwise it take even longer. I was concerned of doing this: what happens if a factor isnt present in the first 1e4 rows? Will it crash? I did a few tests and unknown factor levels are considered as NA and imputed using knn. It’s nice!.
if(FALSE){
tic()
train <- read_fst(here::here("content/post/data/interim/claims_train_1e5.fst")) # , from =1, to = 1e6
rec <- recipes::recipe(Claim_Amount ~ .,
train %>% head(1e5)) %>%
recipes::step_zv(recipes::all_predictors()) %>% # remove variable with all equal values
step_string2factor(all_nominal()) %>% # doesnt lke character columns
step_mutate(
Model_Year = as.factor(Model_Year),
Calendar_Year = as.factor(Calendar_Year)
) %>%
recipes::step_other(recipes::all_nominal() , threshold = 0.01) %>% # combine categories with less than 1% of observation
step_knnimpute(all_predictors()) %>%
recipes::step_dummy(recipes::all_nominal()) %>% # convert to dummy for xgboost use
check_missing(all_predictors()) ## break the bake function if any of the checked columns contains NA value
# Prepare the recipe and use juice/bake to get the d2ata!
trained_rec <- prep(rec)
write_rds(trained_rec, here::here("content/post/data/interim/claims_trained_rec.rds"))
rm(train_1e5)
train <- read_fst(here::here("content/post/data/interim/claims_train.fst"))
train <- bake(trained_rec, new_data = train )
write_fst(train, here::here("content/post/data/interim/claims_baked_train.fst"))
toc() # 190.161 sec elapsed for 1e6 train , 1e4prep # 2017.908 sec elapsed sur train complet (10M) et prep 1e5
rm(list=ls()) # clear memory of object but keep libraries.
invisible(gc())
}if(FALSE){
train <- read_fst(here::here("content/post/data/interim/claims_baked_train.fst"))
train_3e6_split <- rsample::initial_split(train, prop = 0.3)
train_3e6 <- rsample::training(train_3e6_split)
fst::write_fst(train_3e6, here::here("content/post/data/interim/claims_baked_train_3e6.fst"))
rm(train3e6)
rm(train3e6_split)
train_6e6_split <- rsample::initial_split(train, prop = 0.6)
train_6e6 <- rsample::training(train_6e6_split)
fst::write_fst(train_6e6, here::here("content/post/data/interim/claims_baked_train_6e6.fst"))
rm(list=ls()) # clear memory of object but keep libraries.
invisible(gc())
}Train XGBoost
find best number of iterations
if(FALSE){
train <- read_fst( here::here("content/post/data/interim/claims_baked_train_3e6.fst"), from = 1, to = 6e6)
xgtrain <- xgb.DMatrix(as.matrix(train %>% select(-Claim_Amount)),
label = train$Claim_Amount
)
rm(train) # save memory
tic()
params <-list(
booster = "gbtree",
objective = 'reg:tweedie',
eval_metric = "tweedie-nloglik@1.1",
tweedie_variance_power = 1.1,
gamma = 0,
max_depth = 4,
eta = 0.01,
min_child_weight = 3,
subsample = 0.8,
colsample_bytree = 0.8,
tree_method = "hist"
)
xgcv <- xgb.cv(
params = params,
data = xgtrain,
nround = 500,
nfold= 3,
showsd = TRUE,
early_stopping_rounds = 50)
best_iter <- xgcv$best_iteration # 310, 3 folds, 3e6 obs
write_rds(best_iter, here::here("content/post/data/interim/claims_best_iter.rds"))
rm(list=ls()) # clear memory of object but keep libraries.
invisible(gc())
toc()
}if(FALSE){
tic()
train <- read_fst( here::here("content/post/data/interim/claims_baked_train_6e6.fst"), from = 1, to = 6e6)
xgtrain <- xgb.DMatrix(as.matrix(train %>% select(-Claim_Amount)),
label = train$Claim_Amount
)
rm(train) # save memory
best_iter <- read_rds(here::here("content/post/data/interim/claims_best_iter.rds"))
params <-list(
booster = "gbtree",
objective = 'reg:tweedie',
eval_metric = "tweedie-nloglik@1.1",
tweedie_variance_power = 1.1,
gamma = 0,
max_depth = 4,
eta = 0.01,
min_child_weight = 3,
subsample = 0.8,
colsample_bytree = 0.8,
tree_method = "hist"
)
# xgb.cv doesnt ouput any model -- we need a model to predict test dataset
xgmodel <- xgboost::xgb.train(
data = xgtrain,
params = params,
nrounds =best_iter#, # = #310
#nrounds = 50,
#nthread = parallel::detectCores() - 1
)
write_rds(xgmodel, here::here("content/post/data/interim/xgmodel.rds"))
rm(list=ls()) # clear memory of object but keep libraries.
invisible(gc())
toc()
}Apply recipe to test data
if(FALSE){
trained_rec <- read_rds(here::here("content/post/data/interim/claims_trained_rec.rds"))
test <- fst::read_fst( here::here("content/post/data/interim/claims_test.fst"))
test <- bake(trained_rec, new_data = test)
write_fst(test, here::here("content/post/data/interim/claims_baked_test.fst"))
rm(list=ls()) # clear memory of object but keep libraries.
invisible(gc())
}Predict XGBoost
xgmodel <- read_rds(here::here("content/post/data/interim/claims_xgmodel.rds"))
test <- read_fst( here::here("content/post/data/interim/claims_baked_test.fst"))
xgtest <- xgb.DMatrix(as.matrix(test %>% select(-Claim_Amount)),
label = test$Claim_Amount
)
test_w_preds <-
test %>%
mutate(pred_claim_xgboost = predict(xgmodel, xgtest)) %>%
mutate(exposure = 1)
rm(test)
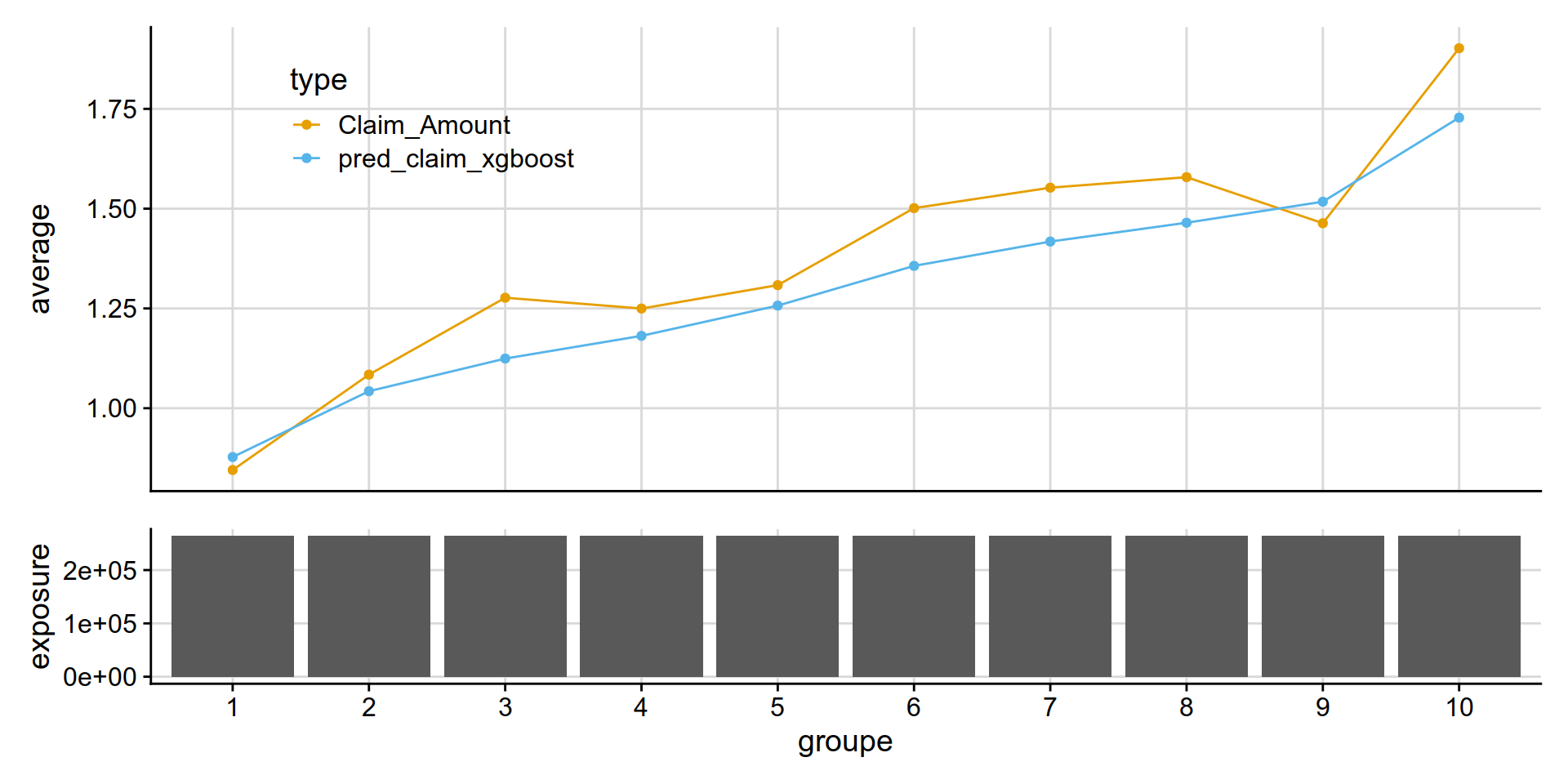
mean(test_w_preds$Claim_Amount) # 1.334191# [1] 1.376243mean(test_w_preds$pred_claim_xgboost) # 1.331216# [1] 1.29676MLmetrics::NormalizedGini(test_w_preds$pred_claim_xgboost, test_w_preds$Claim_Amount) # 0.127837 would have ranked 16 / 102 back in 2012.. not excellent .. but it works# [1] 0.1089691#' @title add_equal_weight_group()
#'
#' @description Cette fonction crée des groupe (quantiles) avec le nombre nombre total d'exposition.
#' @param table data.frame source
#' @param sort_by Variable utilisée pour trier les observations.
#' @param expo Exposition (utilisée pour créer des quantiles de la même taille. Si NULL, l'exposition est égale pour toutes les observations) (Défault = NULL).
#' @param nb Nombre de quantiles crées (défaut = 10)
#' @param group_variable_name Nom de la variable de groupes créée
#' @export
add_equal_weight_group <- function(table, sort_by, expo = NULL, group_variable_name = "groupe", nb = 10) {
sort_by_var <- enquo(sort_by)
groupe_variable_name_var <- enquo(group_variable_name)
if (!(missing(expo))){ # https://stackoverflow.com/questions/48504942/testing-a-function-that-uses-enquo-for-a-null-parameter
expo_var <- enquo(expo)
total <- table %>% pull(!!expo_var) %>% sum
br <- seq(0, total, length.out = nb + 1) %>% head(-1) %>% c(Inf) %>% unique
table %>%
arrange(!!sort_by_var) %>%
mutate(cumExpo = cumsum(!!expo_var)) %>%
mutate(!!group_variable_name := cut(cumExpo, breaks = br, ordered_result = TRUE, include.lowest = TRUE) %>% as.numeric) %>%
select(-cumExpo)
} else {
total <- nrow(table)
br <- seq(0, total, length.out = nb + 1) %>% head(-1) %>% c(Inf) %>% unique
table %>%
arrange(!!sort_by_var) %>%
mutate(cumExpo = row_number()) %>%
mutate(!!group_variable_name := cut(cumExpo, breaks = br, ordered_result = TRUE, include.lowest = TRUE) %>% as.numeric) %>%
select(-cumExpo)
}
}
get_lift_chart_data <- function(
data,
sort_by,
pred,
expo,
obs,
nb = 10) {
pred_var <- enquo(pred)
sort_by_var <- enquo(sort_by)
expo_var <- enquo(expo)
obs_var <- enquo(obs)
pred_name <- quo_name(pred_var)
sort_by_name <- quo_name(sort_by_var)
obs_name <- quo_name(obs_var)
# constitution des buckets de poids égaux
dd <- data %>% add_equal_weight_group(
sort_by = !!sort_by_var,
expo = !!expo_var,
group_variable_name = "groupe",
nb = nb
)
# comparaison sur ces buckets
dd <- full_join(
dd %>%
group_by(groupe) %>%
summarise(
exposure = sum(!!expo_var),
sort_by_moyen = mean(!!sort_by_var),
sort_by_min = min(!!sort_by_var),
sort_by_max = max(!!sort_by_var)
) %>%
ungroup(),
dd %>%
group_by(groupe) %>%
summarise_at(
funs(sum(.) / sum(!!expo_var)),
.vars = vars(!!obs_var, !!pred_var)
) %>%
ungroup,
by = "groupe"
)
# création des labels
dd <- dd %>%
mutate(labs = paste0("[", round(sort_by_min, 2), ", ", round(sort_by_max, 2), "]"))
}
get_lift_chart <- function(data,
sort_by,
pred,
expo,
obs,
nb){
pred_var <- enquo(pred)
sort_by_var <- enquo(sort_by)
expo_var <- enquo(expo)
obs_var <- enquo(obs)
lift_data <- get_lift_chart_data(
data = data,
sort_by = !!sort_by_var,
pred = !!pred_var,
expo = !!expo_var,
obs = !!obs_var,
nb = 10)
p1 <- lift_data %>%
mutate(groupe = as.factor(groupe)) %>%
select(groupe, labs, !!pred_var, !!obs_var) %>%
gather(key = type, value = average, !!pred_var, !!obs_var) %>%
ggplot(aes(x= groupe, y = average, color =type, group = type)) +
geom_line() +
geom_point()+
cowplot::theme_half_open() +
cowplot::background_grid() +
colorblindr::scale_color_OkabeIto( ) +
# scale_y_continuous(
# breaks = scales::pretty_breaks()
# )+
theme(
legend.position = c(0.1, 0.8),
axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank()
)
p2 <- lift_data %>%
mutate(groupe = as.factor(groupe)) %>%
select(groupe, labs, exposure) %>%
ggplot(aes(x= groupe, y = exposure)) +
geom_col() +
cowplot::theme_half_open() +
cowplot::background_grid() +
colorblindr::scale_color_OkabeIto( )# +
# scale_y_continuous(
# expand = c(0,0),
# breaks = scales::breaks_pretty(3)
# )
return(p1 / p2 + plot_layout(heights = c(3, 1)))
}not too bad!
get_lift_chart(
data = test_w_preds,
sort_by= pred_claim_xgboost,
pred = pred_claim_xgboost,
obs = Claim_Amount,
expo = exposure )